Difference between revisions of "HOWTO12 Globus Online cookbook for EGI VOs"
| (134 intermediate revisions by 6 users not shown) | |||
| Line 1: | Line 1: | ||
[[Category: | {{Template:Op menubar}} {{Template:Doc_menubar}} | ||
[[Category:Operations Manuals]] | |||
[[Category:Deprecated]] | |||
{| style="border:1px solid black; background-color:lightgrey; color: black; padding:5px; font-size:140%; width: 90%; margin: auto;" | |||
| style="padding-right: 15px; padding-left: 15px;" | | |||
| | |[[File:Alert.png]] This page is '''Deprecated''' and should no longer be used | ||
|} | |} | ||
{{TOC_right}} | |||
The '''Globus Online cookbook''' describes how the 'SRM type' storage services that are federated into EGI can be used as endpoints of file transfers managed by Globus Online. The "biomed" VO is used as an example in the Cookbook to demonstrate EGI storage usage, but the steps are the same for other EGI VOs as well. | |||
The cookbook was prepared as a guide for EGI Virtual Organisations (VOs) on how to use the Globus Online service that is available at http://www.globusonline.eu. GlobusOnline.eu provides robust and easy to use file transfer capabilities for EGI users. The service manages file transfers for you, monitoring performance, retrying failures, auto-tuning and recovering from faults automatically where possible, and reporting status. | |||
The Cookbook consists of two parts: | |||
#[[Globus Online cookbook for EGI VOs#VO_Managers|For VO Managers ]]: The first part provides step-by-step instructions for VO Managers on how to register SRM storage services in Globus Online in such a way, that these appear as transfer endpoints for VO members. This registration could be performed by any member of a VO, however for most VOs the VO Manager is the most suitable person to complete this step because the VO Manager has sufficient knowledge on storage sites that support the VO and about the BDII information system where detailed information about the storages is recorded. | |||
#[[Globus Online cookbook for EGI VOs#VO_Members|For VO Members (researchers) ]]: The second part provides step-by-step instructions for VO members on how to use VO storage endpoints in Globus Online. This part is relevant for any meber of any EGI VO. The list of EGI VOs and information on joining these VOs is available at http://operations-portal.egi.eu/vo. | |||
''Important note: the http://www.globusonline.eu server is hosted in the US, but the files that the service moves between EGI sites do not leave Europe. The service orchestrates file copies with the GridFTP third party transfer, so files are copied directly between the EGI endpoints. '' | |||
This cookbook has been prepared by the EGI.eu User Community Support Team in consultation with representatives of EGI Operations, storage technology and information system developer groups. Please email any feedback about this Cookbook to the EGI.eu User Community Support Team: ucst@egi.eu. | |||
== VO Managers == | |||
1. Get all endpoints suporting gsiftp for biomed VO (Glue 2.0): | |||
<pre>-bash-3.2$ ldapsearch -LLL -x -h lcg-bdii.cern.ch -p 2170 -b o=glue '(&(objectclass=GLUE2AccessPolicy)(GLUE2PolicyRule=*:biomed))' \ | |||
-bash-3.2$ GLUE2AccessPolicyEndpointForeignKey | perl -p00e 's/\r?\n //g' | grep GLUE2AccessPolicyEndpointForeignKey | sort | uniq | awk '{ print $2 }' |grep -i gsiftp > APEndpoints | |||
</pre> | |||
2. Get all GridFTP (gsiftp) endpoints for biomed VO (Glue 2.0): | |||
<pre>-bash-3.2$ for i in `cat APEndpoints`;do ldapsearch -LLL -x -h lcg-bdii.cern.ch -p 2170 -b o=glue \ | |||
-bash-3.2$ '(&(objectclass=GLUE2Endpoint)(GLUE2EndpointInterfaceName=gsiftp)(GLUE2EndpointID='$i'))' \ | |||
-bash-3.2$ GLUE2EndpointURL GLUE2EndpointImplementationName GLUE2ENdpointImplementationVersion GLUE2EndpointInterfaceName | grep ^GLUE ;echo "-------";done | |||
</pre> | |||
3. Find myproxy server for biomed VO: | |||
<pre>-bash-3.2$ lcg-infosites --is lcg-bdii.cern.ch --vo biomed myproxy | |||
myproxy://px.grid.sara.nl:7512/ | |||
myproxy.cern.ch:7512 | |||
myproxy.usatlas.bnl.gov:7512 | |||
myproxy://cluster6.knu.ac.kr:7512/ | |||
myproxy://grid-mypx.feit.ukim.edu.mk:7512/ | |||
myproxy://grid-px0.desy.de:7512/ | |||
myproxy://grid153.kfki.hu:7512/ | |||
myproxy://gridpx01.ifca.es:7512/ | |||
myproxy://ii.biomed.kiev.ua:7512/ | |||
myproxy://kek2-px.cc.kek.jp:7512/ | |||
myproxy://lcg-px01.icepp.jp:7512/ | |||
myproxy://lcg2proxy.ific.uv.es:7512/ | |||
myproxy://lcgpx01.jinr.ru:7512/ | |||
myproxy://lcgrbp01.gridpp.rl.ac.uk:7512/ | |||
<...> | |||
</pre> | |||
If your VO does not have a dedicated MyPRoxy server, then the catch-all MyProxy server of EGI can be used. This catch-all service is available from CESNET at myproxy.egi.eu.<br> | |||
< | <br> | ||
Extras: example script to obtain GridFTP endpoint information with Glue 2.0. (Command line tools from emi-ui-3.0.0-1.el6.x86_64): | |||
<pre>#!/bin/bash | |||
bdii="top-bdii.cern.ch"; | |||
ldapsearch -LLL -x -H ldap://$bdii:2170 -b o=glue "(&(objectclass=GLUE2AccessPolicy)(GLUE2PolicyRule=*:$1))" GLUE2AccessPolicyEndpointForeignKey | perl -p00e 's/\r?\n //g' | grep GLUE2AccessPolicyEndpointForeignKey |sort | uniq | awk '{ print $2 }' |grep -i gsiftp > APEndpoints | |||
for i in `cat APEndpoints`;do | |||
SEtype=`ldapsearch -LLL -x -H ldap://$bdii:2170 -b o=glue "(&(objectclass=GLUE2Endpoint)(GLUE2EndpointInterfaceName=gsiftp)(GLUE2EndpointID=$i))" GLUE2EndpointImplementationName |grep ^GLUE2| cut -d" " -f2`; | |||
if [ "$SEtype" = "DPM" ]; then | |||
- | SE=`ldapsearch -LLL -x -H ldap://$bdii:2170 -b o=glue "(&(objectclass=GLUE2Endpoint)(GLUE2EndpointInterfaceName=gsiftp)(GLUE2EndpointID=$i))" GLUE2EndpointServiceForeignKey |grep ^GLUE2| cut -d" " -f2`; | ||
fi | |||
if [ "$SEtype" = "dCache" ]; then | |||
( | SE=`ldapsearch -LLL -x -H ldap://$bdii:2170 -b o=glue "(&(objectclass=GLUE2Endpoint)(GLUE2EndpointInterfaceName=gsiftp)(GLUE2EndpointID=$i))" GLUE2EndpointServiceForeignKey |grep ^GLUE2| cut -d" " -f2 | cut -d":" -f2|sed 's/\/data//g'`; | ||
fi | |||
- SE | SRM=`lcg-info --list-service --bdii ldap://$bdii:2170 --vo $1 --query "ServiceType=SRM" --attrs "ServiceEndpoint"|grep $SE | cut -d" " -f3|head -1`; | ||
details=`ldapsearch -LLL -x -H ldap://$bdii:2170 -b o=glue "(&(objectclass=GLUE2Endpoint)(GLUE2EndpointInterfaceName=gsiftp)(GLUE2EndpointID=$i))" GLUE2EndpointURL GLUE2EndpointImplementationName GLUE2ENdpointImplementationVersion | grep ^GLUE`; | |||
VOInfoPath=`lcg-info --list-se --bdii ldap://$bdii:2170 --vo biomed --query "SE=$SE" --attrs "VOInfoPath"|grep VOInfoPath|awk '{print $3}'`; | |||
echo -e "$details\nVOInfoPath: $VOInfoPath\nHost: $SE\nSRM: $SRM\n"; | |||
done; | |||
-bash-3.2$ | </pre> <pre> | ||
-bash-3.2$ ./go.sh biomed | |||
GLUE2EndpointImplementationName: DPM | |||
GLUE2EndpointURL: gsiftp://glite-se.scai.fraunhofer.de:2811 | |||
GLUE2EndpointImplementationVersion: 1.8.8 | |||
VOInfoPath: /dpm/scai.fraunhofer.de/home/biomed | |||
Host: glite-se.scai.fraunhofer.de | |||
SRM: httpg://glite-se.scai.fraunhofer.de:8446/srm/managerv2 | |||
GLUE2EndpointImplementationVersion: 2.6.19 | |||
GLUE2EndpointURL: gsiftp://dcache-door-desy09.desy.de:2811 | |||
GLUE2EndpointImplementationName: dCache | |||
VOInfoPath: /pnfs/desy.de/biomed | |||
Host: dcache-se-desy.desy.de | |||
SRM: httpg://dcache-se-desy.desy.de:8443/srm/managerv2 | |||
<...> | |||
</pre> | |||
Testing access to GridFTP endpoint: | |||
<pre>-bash-3.2$ uberftp glite-se.scai.fraunhofer.de "ls /dpm/scai.fraunhofer.de/home/biomed" | |||
</pre> | |||
<br> | |||
4. Register the endpoints in Globus Online: | |||
a) Go to http://www.globusonline.eu/signup, create an account with your VO name e.g. biomed. (Note that usernames can include only letters and numbers and "_".)<br> (Using the VO name as an account name will ensure that VO members can easily find the endpoints that are available for them.) | |||
b) Upload your ssh public key via globusonline.eu -> "manage identities" | |||
<pre> | c) Now you can manage your account using globusonline.eu client | ||
<pre>-bash-3.2$ ssh biomed@cli.globusonline.eu "help" | |||
</pre> | |||
d) Register the endpoints in the Globus Online service. | |||
Endpoints can be registered through the command line interface or the graphical portal interface of Globus Online. The command line interface allows the association of a default directory with the endpoint and this simplifies the use of the endpoint by VO members. The below example therefore shows the command line tool for endpoint registration. The MyProxy server becomes the default MyProxy, and can be replaced with other MyProxy by VO members during the activation of the endpoint. | |||
<pre>-bash-3.2$ ssh biomed@cli.globusonline.eu "endpoint-add fraunhofer_DE -p gsiftp://glite-se.scai.fraunhofer.de:2811" | |||
-bash-3.2$ ssh biomed@cli.globusonline.eu "endpoint-modify --public fraunhofer_DE" | |||
-bash-3.2$ ssh biomed@cli.globusonline.eu "endpoint-modify --myproxy-server=px.grid.sara.nl fraunhofer_DE" | |||
-bash-3.2$ ssh biomed@cli.globusonline.eu "endpoint-modify --default-directory=/dpm/scai.fraunhofer.de/home/biomed fraunhofer_DE" | |||
-bash-3.2$ ssh biomed@cli.globusonline.eu "endpoint-list -v fraunhofer_DE" | |||
Name : biomed#fraunhofer_DE | |||
Host(s) : gsiftp://glite-se.scai.fraunhofer.de:2811 | |||
Subject(s) : | |||
Target Endpoint : n/a | |||
Default Directory : /dpm/scai.fraunhofer.de/home/biomed | |||
Force Encrypted Transfer: No | |||
Disable Verify : No | |||
MyProxy Server : px.grid.sara.nl | |||
MyProxy DN : n/a | |||
MyProxy OAuth Server : n/a | |||
Credential Status : EXPIRED | |||
Credential Expires : | |||
Credential Subject : | |||
</pre> | |||
<br> | |||
== VO Members == | |||
'''1. Generate a VOMS proxy and upload it into a MyProxy server.''' ' | |||
There are two ways to do this: | |||
a). With a graphical tool, such as GSISSH-Term. | |||
OR | |||
b). With the command line tools of the User Interface machine of your VO. | |||
The usage of GSISSH-Term for proxy management is explained on a [[MyProxy tool GUI|dedicated page]]. A usage of the command line tools is detailed below. | |||
<br> | |||
Generating a VOMS proxy with the command line tools (You should have these installed on the User Interface machine of your VO): | |||
<pre>-bash-3.2$ voms-proxy-init --voms biomed | |||
Enter GRID pass phrase for this identity: | |||
Contacting cclcgvomsli01.in2p3.fr:15000 [/O=GRID-FR/C=FR/O=CNRS/OU=CC-IN2P3/CN=cclcgvomsli01.in2p3.fr] "biomed"... | |||
Remote VOMS server contacted succesfully. | |||
Created proxy in /tmp/x509up_u507. | |||
Your proxy is valid until Fri May 24 04:12:03 CEST 2013 | |||
</pre> | |||
'''VOMS proxy lifetime: by default voms proxy extension is generated for 12hours, some voms servers within EGI allow to have lifetime for 1 week (168hours), some allow up to 24hours, please consult your VO manager.''' | |||
<br> Upload VOMS proxy to a MyProxy server with the command line tool. Note: If your VO does not have any MyProxy server, then you can use the EGI catch-all MyProxy server. [[EGI Myproxy with OAuth|Further information about the EGI catch-all MyProxy server]]. | |||
<pre>-bash-3.2$ myproxy-init -l <CHOOSE ANY USERNAME> -s px.grid.sara.nl | |||
Your identity: /O=dutchgrid/O=users/O=egi/CN=Karolis Eigelis | Your identity: /O=dutchgrid/O=users/O=egi/CN=Karolis Eigelis | ||
Enter GRID pass phrase for this identity: | |||
Creating proxy ........................................................................ Done | |||
Proxy Verify OK | |||
Your proxy is valid until: Thu May 30 16:15:47 2013 | |||
Enter MyProxy pass phrase: <YOU NEW PASSWORD TO BE USED LATER AT GLOBUSONLINE> | |||
Verifying - Enter MyProxy pass phrase: <YOU NEW PASSWORD TO BE USED LATER AT GLOBUSONLINE> | |||
A proxy valid for 168 hours (7.0 days) for user <YOUR USERNAME SPECIFIED WITH -l WITHIN THE COMMAND> now exists on px.grid.sara.nl. | |||
</pre> | |||
'''ATTENTION:''' The password entered for MyProxy will be used to authenticate the user via GlobusOnline.eu and the user name is the one which is chosen by you and provided with "-l" argument.<br> | |||
</pre> | |||
- | |||
< | |||
<br> | |||
'''2. Perform file transfers''' | |||
a) Go to http://www.globusonline.eu/signup and create an account. (The account can be later associated with your EGI Single Sign-On account ([http://egi.eu/sso EGI SSO]). '''Attention:''' login with the EGI Single Sign On account is possible only if you have a valid VOMS proxy in the EGI catch-all MyProxy server.)<br> | |||
b) Go to "Start Transfer"<br> | |||
c) Find the transfer endpoints that are available for your VO by searching for your VO name in the endpoint field e.g. biomed<br> | |||
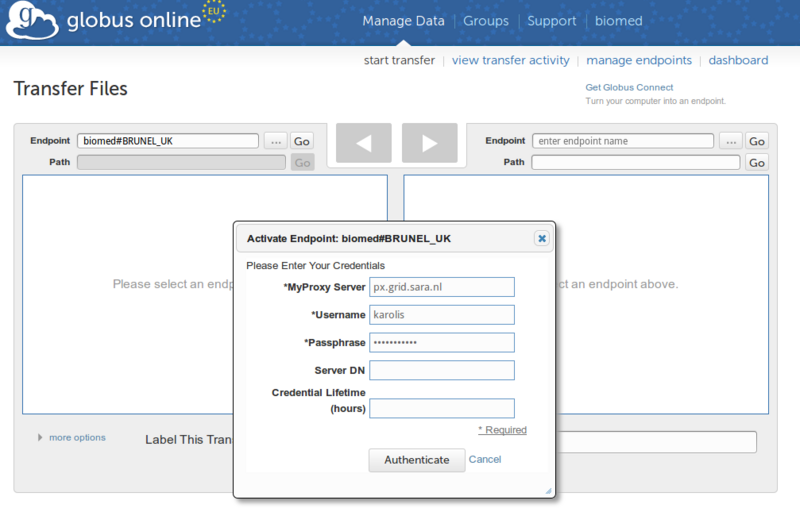
[[Image:Gobiomed.png|thumb|none|800px]]<br> | |||
'''IMPORTANT TO KNOW:''' you may use field "Credential Lifetime (hours)" and enter 168hours - 1 week of proxy lifetime. (Default is 12hours). What is important to understand is that GlobusOnline.eu will retrieve the instance of your proxy from MyProxy server and will activate the endpoint for 168hours - within the GlobusOnline.eu interface you will see that endpoint is activated for 168hours, but this might not be true because your voms proxy lifetime depends on your VOMS server, where you should consult your VO Manager. Some EGI voms servers allow up to 24hours only of the voms proxy lifetime to be. | |||
e) Click "Authenticate"<br> | |||
What you can do with the endpoints? <br> | |||
*Transfer files from your laptop using GlobusConnect client to a endpoint using Globus Online.<br> | |||
*Transfer files from an endpoint to your laptop using GlobusConnect client and Globus Online.<br> | |||
*Transfer files from an endpoint to another endpoint using Globus Online.<br> | |||
Please consult with the Globus Online documentations that are available at http://www.globusonline.eu. | |||
== Additional materials == | |||
The tests that have been carried out while creating this Cookbook are available at [[GO testing]] | |||
Latest revision as of 14:48, 10 September 2021
| Main | EGI.eu operations services | Support | Documentation | Tools | Activities | Performance | Technology | Catch-all Services | Resource Allocation | Security |
| Documentation menu: | Home • | Manuals • | Procedures • | Training • | Other • | Contact ► | For: | VO managers • | Administrators |
The Globus Online cookbook describes how the 'SRM type' storage services that are federated into EGI can be used as endpoints of file transfers managed by Globus Online. The "biomed" VO is used as an example in the Cookbook to demonstrate EGI storage usage, but the steps are the same for other EGI VOs as well.
The cookbook was prepared as a guide for EGI Virtual Organisations (VOs) on how to use the Globus Online service that is available at http://www.globusonline.eu. GlobusOnline.eu provides robust and easy to use file transfer capabilities for EGI users. The service manages file transfers for you, monitoring performance, retrying failures, auto-tuning and recovering from faults automatically where possible, and reporting status.
The Cookbook consists of two parts:
- For VO Managers : The first part provides step-by-step instructions for VO Managers on how to register SRM storage services in Globus Online in such a way, that these appear as transfer endpoints for VO members. This registration could be performed by any member of a VO, however for most VOs the VO Manager is the most suitable person to complete this step because the VO Manager has sufficient knowledge on storage sites that support the VO and about the BDII information system where detailed information about the storages is recorded.
- For VO Members (researchers) : The second part provides step-by-step instructions for VO members on how to use VO storage endpoints in Globus Online. This part is relevant for any meber of any EGI VO. The list of EGI VOs and information on joining these VOs is available at http://operations-portal.egi.eu/vo.
Important note: the http://www.globusonline.eu server is hosted in the US, but the files that the service moves between EGI sites do not leave Europe. The service orchestrates file copies with the GridFTP third party transfer, so files are copied directly between the EGI endpoints.
This cookbook has been prepared by the EGI.eu User Community Support Team in consultation with representatives of EGI Operations, storage technology and information system developer groups. Please email any feedback about this Cookbook to the EGI.eu User Community Support Team: ucst@egi.eu.
VO Managers
1. Get all endpoints suporting gsiftp for biomed VO (Glue 2.0):
-bash-3.2$ ldapsearch -LLL -x -h lcg-bdii.cern.ch -p 2170 -b o=glue '(&(objectclass=GLUE2AccessPolicy)(GLUE2PolicyRule=*:biomed))' \
-bash-3.2$ GLUE2AccessPolicyEndpointForeignKey | perl -p00e 's/\r?\n //g' | grep GLUE2AccessPolicyEndpointForeignKey | sort | uniq | awk '{ print $2 }' |grep -i gsiftp > APEndpoints
2. Get all GridFTP (gsiftp) endpoints for biomed VO (Glue 2.0):
-bash-3.2$ for i in `cat APEndpoints`;do ldapsearch -LLL -x -h lcg-bdii.cern.ch -p 2170 -b o=glue \ -bash-3.2$ '(&(objectclass=GLUE2Endpoint)(GLUE2EndpointInterfaceName=gsiftp)(GLUE2EndpointID='$i'))' \ -bash-3.2$ GLUE2EndpointURL GLUE2EndpointImplementationName GLUE2ENdpointImplementationVersion GLUE2EndpointInterfaceName | grep ^GLUE ;echo "-------";done
3. Find myproxy server for biomed VO:
-bash-3.2$ lcg-infosites --is lcg-bdii.cern.ch --vo biomed myproxy myproxy://px.grid.sara.nl:7512/ myproxy.cern.ch:7512 myproxy.usatlas.bnl.gov:7512 myproxy://cluster6.knu.ac.kr:7512/ myproxy://grid-mypx.feit.ukim.edu.mk:7512/ myproxy://grid-px0.desy.de:7512/ myproxy://grid153.kfki.hu:7512/ myproxy://gridpx01.ifca.es:7512/ myproxy://ii.biomed.kiev.ua:7512/ myproxy://kek2-px.cc.kek.jp:7512/ myproxy://lcg-px01.icepp.jp:7512/ myproxy://lcg2proxy.ific.uv.es:7512/ myproxy://lcgpx01.jinr.ru:7512/ myproxy://lcgrbp01.gridpp.rl.ac.uk:7512/ <...>
If your VO does not have a dedicated MyPRoxy server, then the catch-all MyProxy server of EGI can be used. This catch-all service is available from CESNET at myproxy.egi.eu.
Extras: example script to obtain GridFTP endpoint information with Glue 2.0. (Command line tools from emi-ui-3.0.0-1.el6.x86_64):
#!/bin/bash
bdii="top-bdii.cern.ch";
ldapsearch -LLL -x -H ldap://$bdii:2170 -b o=glue "(&(objectclass=GLUE2AccessPolicy)(GLUE2PolicyRule=*:$1))" GLUE2AccessPolicyEndpointForeignKey | perl -p00e 's/\r?\n //g' | grep GLUE2AccessPolicyEndpointForeignKey |sort | uniq | awk '{ print $2 }' |grep -i gsiftp > APEndpoints
for i in `cat APEndpoints`;do
SEtype=`ldapsearch -LLL -x -H ldap://$bdii:2170 -b o=glue "(&(objectclass=GLUE2Endpoint)(GLUE2EndpointInterfaceName=gsiftp)(GLUE2EndpointID=$i))" GLUE2EndpointImplementationName |grep ^GLUE2| cut -d" " -f2`;
if [ "$SEtype" = "DPM" ]; then
SE=`ldapsearch -LLL -x -H ldap://$bdii:2170 -b o=glue "(&(objectclass=GLUE2Endpoint)(GLUE2EndpointInterfaceName=gsiftp)(GLUE2EndpointID=$i))" GLUE2EndpointServiceForeignKey |grep ^GLUE2| cut -d" " -f2`;
fi
if [ "$SEtype" = "dCache" ]; then
SE=`ldapsearch -LLL -x -H ldap://$bdii:2170 -b o=glue "(&(objectclass=GLUE2Endpoint)(GLUE2EndpointInterfaceName=gsiftp)(GLUE2EndpointID=$i))" GLUE2EndpointServiceForeignKey |grep ^GLUE2| cut -d" " -f2 | cut -d":" -f2|sed 's/\/data//g'`;
fi
SRM=`lcg-info --list-service --bdii ldap://$bdii:2170 --vo $1 --query "ServiceType=SRM" --attrs "ServiceEndpoint"|grep $SE | cut -d" " -f3|head -1`;
details=`ldapsearch -LLL -x -H ldap://$bdii:2170 -b o=glue "(&(objectclass=GLUE2Endpoint)(GLUE2EndpointInterfaceName=gsiftp)(GLUE2EndpointID=$i))" GLUE2EndpointURL GLUE2EndpointImplementationName GLUE2ENdpointImplementationVersion | grep ^GLUE`;
VOInfoPath=`lcg-info --list-se --bdii ldap://$bdii:2170 --vo biomed --query "SE=$SE" --attrs "VOInfoPath"|grep VOInfoPath|awk '{print $3}'`;
echo -e "$details\nVOInfoPath: $VOInfoPath\nHost: $SE\nSRM: $SRM\n";
done;
-bash-3.2$ ./go.sh biomed
GLUE2EndpointImplementationName: DPM GLUE2EndpointURL: gsiftp://glite-se.scai.fraunhofer.de:2811 GLUE2EndpointImplementationVersion: 1.8.8 VOInfoPath: /dpm/scai.fraunhofer.de/home/biomed Host: glite-se.scai.fraunhofer.de SRM: httpg://glite-se.scai.fraunhofer.de:8446/srm/managerv2
GLUE2EndpointImplementationVersion: 2.6.19 GLUE2EndpointURL: gsiftp://dcache-door-desy09.desy.de:2811 GLUE2EndpointImplementationName: dCache VOInfoPath: /pnfs/desy.de/biomed Host: dcache-se-desy.desy.de SRM: httpg://dcache-se-desy.desy.de:8443/srm/managerv2
<...>
Testing access to GridFTP endpoint:
-bash-3.2$ uberftp glite-se.scai.fraunhofer.de "ls /dpm/scai.fraunhofer.de/home/biomed"
4. Register the endpoints in Globus Online:
a) Go to http://www.globusonline.eu/signup, create an account with your VO name e.g. biomed. (Note that usernames can include only letters and numbers and "_".)
(Using the VO name as an account name will ensure that VO members can easily find the endpoints that are available for them.)
b) Upload your ssh public key via globusonline.eu -> "manage identities"
c) Now you can manage your account using globusonline.eu client
-bash-3.2$ ssh biomed@cli.globusonline.eu "help"
d) Register the endpoints in the Globus Online service.
Endpoints can be registered through the command line interface or the graphical portal interface of Globus Online. The command line interface allows the association of a default directory with the endpoint and this simplifies the use of the endpoint by VO members. The below example therefore shows the command line tool for endpoint registration. The MyProxy server becomes the default MyProxy, and can be replaced with other MyProxy by VO members during the activation of the endpoint.
-bash-3.2$ ssh biomed@cli.globusonline.eu "endpoint-add fraunhofer_DE -p gsiftp://glite-se.scai.fraunhofer.de:2811" -bash-3.2$ ssh biomed@cli.globusonline.eu "endpoint-modify --public fraunhofer_DE" -bash-3.2$ ssh biomed@cli.globusonline.eu "endpoint-modify --myproxy-server=px.grid.sara.nl fraunhofer_DE" -bash-3.2$ ssh biomed@cli.globusonline.eu "endpoint-modify --default-directory=/dpm/scai.fraunhofer.de/home/biomed fraunhofer_DE" -bash-3.2$ ssh biomed@cli.globusonline.eu "endpoint-list -v fraunhofer_DE" Name : biomed#fraunhofer_DE Host(s) : gsiftp://glite-se.scai.fraunhofer.de:2811 Subject(s) : Target Endpoint : n/a Default Directory : /dpm/scai.fraunhofer.de/home/biomed Force Encrypted Transfer: No Disable Verify : No MyProxy Server : px.grid.sara.nl MyProxy DN : n/a MyProxy OAuth Server : n/a Credential Status : EXPIRED Credential Expires : Credential Subject :
VO Members
1. Generate a VOMS proxy and upload it into a MyProxy server. '
There are two ways to do this:
a). With a graphical tool, such as GSISSH-Term.
OR
b). With the command line tools of the User Interface machine of your VO.
The usage of GSISSH-Term for proxy management is explained on a dedicated page. A usage of the command line tools is detailed below.
Generating a VOMS proxy with the command line tools (You should have these installed on the User Interface machine of your VO):
-bash-3.2$ voms-proxy-init --voms biomed Enter GRID pass phrase for this identity: Contacting cclcgvomsli01.in2p3.fr:15000 [/O=GRID-FR/C=FR/O=CNRS/OU=CC-IN2P3/CN=cclcgvomsli01.in2p3.fr] "biomed"... Remote VOMS server contacted succesfully. Created proxy in /tmp/x509up_u507. Your proxy is valid until Fri May 24 04:12:03 CEST 2013
VOMS proxy lifetime: by default voms proxy extension is generated for 12hours, some voms servers within EGI allow to have lifetime for 1 week (168hours), some allow up to 24hours, please consult your VO manager.
Upload VOMS proxy to a MyProxy server with the command line tool. Note: If your VO does not have any MyProxy server, then you can use the EGI catch-all MyProxy server. Further information about the EGI catch-all MyProxy server.
-bash-3.2$ myproxy-init -l <CHOOSE ANY USERNAME> -s px.grid.sara.nl Your identity: /O=dutchgrid/O=users/O=egi/CN=Karolis Eigelis Enter GRID pass phrase for this identity: Creating proxy ........................................................................ Done Proxy Verify OK Your proxy is valid until: Thu May 30 16:15:47 2013 Enter MyProxy pass phrase: <YOU NEW PASSWORD TO BE USED LATER AT GLOBUSONLINE> Verifying - Enter MyProxy pass phrase: <YOU NEW PASSWORD TO BE USED LATER AT GLOBUSONLINE> A proxy valid for 168 hours (7.0 days) for user <YOUR USERNAME SPECIFIED WITH -l WITHIN THE COMMAND> now exists on px.grid.sara.nl.
ATTENTION: The password entered for MyProxy will be used to authenticate the user via GlobusOnline.eu and the user name is the one which is chosen by you and provided with "-l" argument.
2. Perform file transfers
a) Go to http://www.globusonline.eu/signup and create an account. (The account can be later associated with your EGI Single Sign-On account (EGI SSO). Attention: login with the EGI Single Sign On account is possible only if you have a valid VOMS proxy in the EGI catch-all MyProxy server.)
b) Go to "Start Transfer"
c) Find the transfer endpoints that are available for your VO by searching for your VO name in the endpoint field e.g. biomed
IMPORTANT TO KNOW: you may use field "Credential Lifetime (hours)" and enter 168hours - 1 week of proxy lifetime. (Default is 12hours). What is important to understand is that GlobusOnline.eu will retrieve the instance of your proxy from MyProxy server and will activate the endpoint for 168hours - within the GlobusOnline.eu interface you will see that endpoint is activated for 168hours, but this might not be true because your voms proxy lifetime depends on your VOMS server, where you should consult your VO Manager. Some EGI voms servers allow up to 24hours only of the voms proxy lifetime to be.
e) Click "Authenticate"
What you can do with the endpoints?
- Transfer files from your laptop using GlobusConnect client to a endpoint using Globus Online.
- Transfer files from an endpoint to your laptop using GlobusConnect client and Globus Online.
- Transfer files from an endpoint to another endpoint using Globus Online.
Please consult with the Globus Online documentations that are available at http://www.globusonline.eu.
Additional materials
The tests that have been carried out while creating this Cookbook are available at GO testing