Difference between revisions of "HOWTO12 Globus Online cookbook for EGI VOs"
| Line 14: | Line 14: | ||
</p><p><b>This cookbook has been prepared by the EGI.eu User Community Support Team in consultation with representatives of EGI Operations, storage and information system | </p><p><b>This cookbook has been prepared by the EGI.eu User Community Support Team in consultation with representatives of EGI Operations, storage and information system developer groups. Please email any feedback about this Cookbook to the EGI.eu User Community Support Team: ucst@egi.eu.</b></p> | ||
<h2>VO Managers </h2> | <h2>VO Managers </h2> | ||
Revision as of 15:25, 24 June 2013
About
This page provides guidance for Virtual Organisations (VO) of the European Grid Infrastructure (EGI) on how to use the Globus Online service that is available at http://www.globusonline.eu. The service provides robust and easy to use file transfer capabilities for EGI users. Globus Online manages file transfers for you, monitoring performance, retrying failures, auto-tuning and recovering from faults automatically where possible, and reporting status. The Cookbook describes how SRM storage service in EGI could be used as endpoints for file transfers in Globus Online. The "biomed" VO is used as an example in the Cookbook to demonstrate EGI storage usage.
The Cookbook consists of two parts:
- VO Managers: The first part provides step-by-step instructions for VO Managers on how to register SRM storage services in Globus Online in such a way, that these appear as transfer endpoints for VO members. These steps could be performed by any member of a VO, and we assume that for most VOs the VO Manager is the most suitable person to complete the steps. VO Managers have sufficient knowledge on storage sites that support the VO and about the BDII information system where detailed information about these storages is recorded.
- VO Members: The second part provides step-by-step instructions for VO members on how to use storage endpoints in Globus Online. These steps can be carried out by any EGI VO member. The list of EGI VOs and information on joining these VOs is available at http://operations-portal.egi.eu/vo.
This cookbook has been prepared by the EGI.eu User Community Support Team in consultation with representatives of EGI Operations, storage and information system developer groups. Please email any feedback about this Cookbook to the EGI.eu User Community Support Team: ucst@egi.eu.
VO Managers
1. Get all storage managers (Glue 2.0):
-bash-3.2$ ldapsearch -x -h lcg-bdii.cern.ch -p 2170 -b o=glue '(&(objectclass=GLUE2StorageService))' |perl -p00e 's/\r?\n //g'|grep ^dn:|cut -d" " -f2 > GLUE2StorageServiceDN
2. Filter out only storage managers for biomed VO (Glue 2.0):
-bash-3.2$ for i in `cat GLUE2StorageServiceDN`;do ldapsearch -LLL -x -H ldap://lcg-bdii.cern.ch:2170 -x -b "$i" "(&(objectClass=GLUE2AccessPolicy)(GLUE2PolicyRule=*:biomed))" GLUE2PolicyRule| grep -q GLUE2PolicyRule && echo $i;done > GLUE2AccessPolicyDN
3. Get all GridFTP endpoints for biomed VO (Glue 2.0):
-bash-3.2$ for i in `cat GLUE2AccessPolicyDN`; do ldapsearch -LLL -x -H ldap://lcg-bdii.cern.ch:2170 -x -b "$i" "(&(objectClass=GLUE2Endpoint)(GLUE2EndpointInterfaceName=gsiftp))" GLUE2EndpointURL | grep ^GLUE |tail -n 1; echo StorageServiceID: $i|cut -d"," -f1|sed 's/GLUE2ServiceID=//g';echo "-------";done GLUE2EndpointURL: gsiftp://gaeds020.ciemat.es:2811 StorageServiceID: glue:srm.ciemat.es/data ------- GLUE2EndpointURL: gsiftp://se001.ipp.acad.bg:2811 StorageServiceID: glue:se001.ipp.acad.bg/data ------- GLUE2EndpointURL: gsiftp://juliet.zih.tu-dresden.de:2811 StorageServiceID: glue:ophelia.zih.tu-dresden.de/data ------- GLUE2EndpointURL: gsiftp://sedsk58.grid.hep.ph.ic.ac.uk:2811 StorageServiceID: glue:gfe02.grid.hep.ph.ic.ac.uk/data -------
ATTENTION: you might not get all the GridFTP endpoints, because at the moment only dCache is publishing them to information system. Currently DPM is working on a feature to publish gridftp endpoints (REF: https://its.cern.ch/jira/browse/LCGDM-1083), so for the time being please pay attention to Additional steps for finding the GridFTP endpoints for DPM and StoRM (Glue 1.3):.
4. Find myproxy server for biomed VO:
-bash-3.2$ lcg-infosites --is lcg-bdii.cern.ch --vo biomed myproxy myproxy.cern.ch:7512 myproxy.usatlas.bnl.gov:7512 myproxy://cluster6.knu.ac.kr:7512/ myproxy://grid-mypx.feit.ukim.edu.mk:7512/ myproxy://grid-px0.desy.de:7512/ myproxy://grid153.kfki.hu:7512/ myproxy://gridpx01.ifca.es:7512/ myproxy://ii.biomed.kiev.ua:7512/ myproxy://kek2-px.cc.kek.jp:7512/ myproxy://lcg-px01.icepp.jp:7512/ myproxy://lcg2proxy.ific.uv.es:7512/ myproxy://lcgpx01.jinr.ru:7512/ myproxy://lcgrbp01.gridpp.rl.ac.uk:7512/ <...>
Additional steps for finding the GridFTP endpoints for DPM and StoRM (Glue 1.3):
Extra step 1. Example script to get the needed info (Glue 1.3. Command line tools from emi-ui-3.0.0-1.el6.x86_64):
#!/bin/bash
bdii="lcg-bdii.cern.ch";
lcg-infosites --is $bdii --vo $1 se |awk '{print $4}'|sort|uniq > $1
for host in `cat $1`; do
ServiceEndpoint=`lcg-info --list-service --bdii ldap://lcg-bdii.cern.ch:2170 --vo $1 --query "ServiceType=SRM" --attrs "ServiceEndpoint"|grep $host|cut -d" " -f3|head -1;`
VOInfoPath=`lcg-info --list-se --bdii ldap://$bdii:2170 --vo $1 --query "SE=$host" --attrs "VOInfoPath"|grep VOInfoPath|awk '{print $3}'`;
gsiftp=`ldapsearch -LLL -x -H ldap://$bdii:2170 -x -b o=grid "(&(GlueSEAccessProtocolType=gsiftp)(GlueChunkKey=GlueSEUniqueID=$host))" GlueSEAccessProtocolEndpoint | grep ^Glue |awk '{print $2}'| tail -1`;
details=`ldapsearch -LLL -h lcg-bdii.cern.ch -p 2170 -x -b o=grid "(&(objectClass=GlueSE)(GlueSEUniqueID=$host))" GlueSEImplementationName GlueSEImplementationVersion|grep ^Glue`;
echo -e "Host: $host\nSRMServiceEndpoint: $ServiceEndpoint\nVOInfoPath: $VOInfoPath\nGlueSEAccessProtocolEndpoint: $gsiftp\n$details\n";
done;
-bash-3.2$ ./go.sh biomed Host: dcache-se-desy.desy.de SRMServiceEndpoint: httpg://dcache-se-desy.desy.de:8443/srm/managerv2 VOInfoPath: /pnfs/desy.de/biomed GlueSEAccessProtocolEndpoint: gsiftp://dcache-door-desy09.desy.de:2811 GlueSEImplementationVersion: 1.9.12-12 (ns=Chimera) GlueSEImplementationName: dCache Host: dc2-grid-64.brunel.ac.uk SRMServiceEndpoint: httpg://dc2-grid-64.brunel.ac.uk:8446/srm/managerv2 VOInfoPath: /dpm/brunel.ac.uk/home/biomed GlueSEAccessProtocolEndpoint: GlueSEImplementationVersion: 1.8.6 GlueSEImplementationName: DPM Host: grid2.fe.infn.it SRMServiceEndpoint: httpg://grid2.fe.infn.it:8444/srm/managerv2 VOInfoPath: /biomed GlueSEAccessProtocolEndpoint: GlueSEImplementationVersion: 1.10.0 GlueSEImplementationName: StoRM <...>
Extra step 2. Finding GridFTP endpoint for DPM or StoRM:
-bash-3.2$ touch test -bash-3.2$ lcg-cp file://$PWD/test srm://dc2-grid-64.brunel.ac.uk:8446/dpm/brunel.ac.uk/home/biomed/test -bash-3.2$ lcg-gt srm://dc2-grid-64.brunel.ac.uk:8446/dpm/brunel.ac.uk/home/biomed/test gsiftp gsiftp://dc2-grid-pool-a4-02.brunel.ac.uk/dc2-grid-pool-a4-02.brunel.ac.uk:/data2/dpmfs/biomed/2013-05-23/test.29428758.0 d4f44efc-b0c2-4ab2-ba0d-1c80f758e25b -bash-3.2$ lcg-del -l srm://dc2-grid-64.brunel.ac.uk:8446/dpm/brunel.ac.uk/home/biomed/test
Making sure that GridFTP endpoint is accessible:
-bash-3.2$ uberftp dc2-grid-pool-a4-02.brunel.ac.uk "ls /dpm/brunel.ac.uk/home/biomed"
5. Go to http://www.globusonline.eu/
a) Create an account with your VO name e.g. biomed. (Note that usernames can include only letters and numbers and "_".)
b) Upload your ssh public key via globusonline.eu -> "manage identities"
c) Now you can manage your account using globusonline.eu client
-bash-3.2$ ssh biomed@cli.globusonline.eu "help"
d) Lets create the endpoint with the all needed information we have
-bash-3.2$ ssh biomed@cli.globusonline.eu "endpoint-add BRUNEL_UK -p gsiftp://dc2-grid-64.brunel.ac.uk:2811" -bash-3.2$ ssh biomed@cli.globusonline.eu "endpoint-modify --public BRUNEL_UK" -bash-3.2$ ssh biomed@cli.globusonline.eu "endpoint-modify --myproxy-server=px.grid.sara.nl BRUNEL_UK" -bash-3.2$ ssh biomed@cli.globusonline.eu "endpoint-modify --default-directory=/dpm/brunel.ac.uk/home/biomed/ BRUNEL_UK" -bash-3.2$ ssh biomed@cli.globusonline.eu "endpoint-list -v BRUNEL_UK" Name : biomed#BRUNEL_UK Host(s) : gsiftp://dc2-grid-64.brunel.ac.uk:2811 Subject(s) : Target Endpoint : n/a Default Directory : /dpm/brunel.ac.uk/home/biomed/ Force Encrypted Transfer: No Disable Verify : No MyProxy Server : px.grid.sara.nl MyProxy DN : n/a MyProxy OAuth Server : n/a Credential Status : EXPIRED Credential Expires : Credential Subject :
VO Members
1. Generating credentials.
Generate VOMS proxy:
-bash-3.2$ voms-proxy-init --voms biomed Enter GRID pass phrase for this identity: Contacting cclcgvomsli01.in2p3.fr:15000 [/O=GRID-FR/C=FR/O=CNRS/OU=CC-IN2P3/CN=cclcgvomsli01.in2p3.fr] "biomed"... Remote VOMS server contacted succesfully. Created proxy in /tmp/x509up_u507. Your proxy is valid until Fri May 24 04:12:03 CEST 2013
Upload VOMS proxy to MyProxy.
-bash-3.2$ myproxy-init -s px.grid.sara.nl Your identity: /O=dutchgrid/O=users/O=egi/CN=Karolis Eigelis Enter GRID pass phrase for this identity: Creating proxy ........................................................................ Done Proxy Verify OK Your proxy is valid until: Thu May 30 16:15:47 2013 Enter MyProxy pass phrase: <YOU NEW PASSWORD TO BE USED LATER AT GLOBUSONLINE> Verifying - Enter MyProxy pass phrase: <YOU NEW PASSWORD TO BE USED LATER AT GLOBUSONLINE> A proxy valid for 168 hours (7.0 days) for user karolis now exists on px.grid.sara.nl.
ATTENTION: The password you enter for MyProxy will be used to authenticate you via globusonline.eu and the user name is the one which is given by myproxy server, in this case - "karolis".
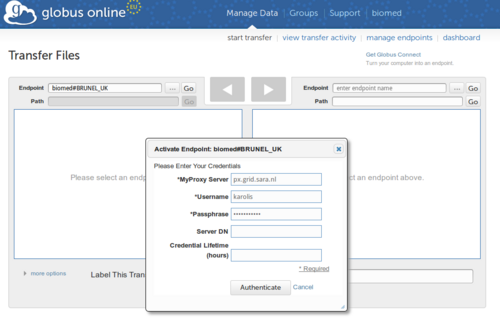
2. Go to to http://www.globusonline.eu/
a) Create account for yourself
b) Go to "Start Transfer"
c) Search for your VO name in the endpoint field e.g. biomed
e) Click "Authenticate"
That is it. Now using Globus Online you can access Storage Element in EGI which is used also by SRM and FTS.
What you can do ?
- Transfer files from your laptop using GlobusConnect client to SRM storage endpoint in EGI using GO.
- Transfer files from SRM endpoint in EGI using GO to your laptop using GlobusConnect client.
- Transfer files from one SRM endpoint in EGI to another SRM endpoint in EGI or to any other Infrastructure using GO and receive nice notifications by email
Additional materials
Tests have been carried out while creating this Cookbook: GO_testing